Topics¶
- Classification: binary, multi-class
- Logistic Regression
- Naïve Bayes Classification
- K-nearest Neighbours
- Support Vector Machines
Classification¶
Given a sample with $n$ independent features
$X^i = [x^i_1, x^i_2, ..., x^i_n]$
Predict the probability $P(y)$ that this sample belongs to a class $y$
i.e. we "classify" the sample as belonging to $y$
Algorithms for Classification¶
- Logistic Regression*
- Naive Bayes*
- K-nearest Neighbours*
- Support Vector Machines*
- Decision Trees and Forests
- Neural Networks
- etc
[* Covered today]
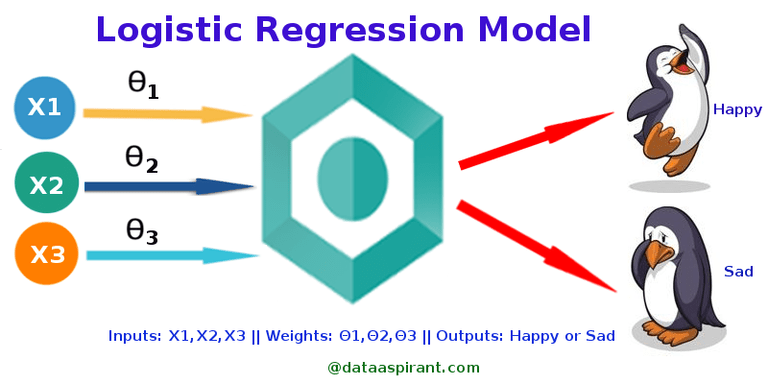
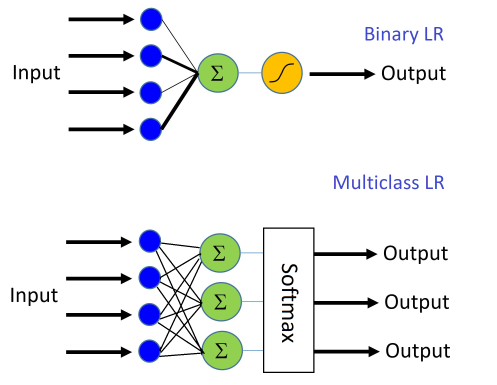
Logistic Regression¶
Linear Regression + either Activation or Softmax
Activation: Binary Classification
Softmax: Multi-class Classification
Logistic Sigmoid¶
Converts the output to a value between 0 and 1
- 1 can mean True, Happy, ...
- 0 can mean False, Sad, ...
$$\sigma(x) = \frac{1}{1+exp(-x)}$$
# Credits: https://ilparle.com/2017/04/21/plot-a-simple-sigmoid-function/
import numpy as np
import matplotlib.pyplot as plt
x = np.arange(-8, 8, 0.1)
sigmoid = 1 / (1 + np.exp(-x))
fig, ax = plt.subplots()
ax.plot(x, sigmoid)
ax.set(xlabel = 'x', ylabel = 'sigmoid')
ax.grid()
plt.show()
Softmax¶
Softmax:
- Converts multiple outputs to a percentage distribution between 0 and 1
- Percentage distribution: numbers all add up to 1 (100%)
- Outputs: 0.7 happy, 0.2 depressed, 0.1 unknown
Example: [1, 2, 3, 4, 1, 2, 3]
Result: [0.024, 0.064, 0.175, 0.475, 0.024, 0.064, 0.175]
# Credits: https://en.wikipedia.org/wiki/Softmax_function
import numpy as np
z = [1.0, 2.0, 3.0, 4.0, 1.0, 2.0, 3.0]
softmax = lambda x : np.exp(x)/np.sum(np.exp(x))
softmax(z)
(image: CNTK)
# Iris flower dataset
# https://archive.ics.uci.edu/ml/datasets/iris
import sklearn
from sklearn import datasets
from sklearn.model_selection import train_test_split
iris = datasets.load_iris()
X = iris.data
y = iris.target
# Add noisy features to make the problem harder
random_state = np.random.RandomState(0)
n_samples, n_features = X.shape
X = np.c_[X, random_state.randn(n_samples, 200 * n_features)]
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.5, random_state=99)
print('First 5 training data:', X_train[:5])
print('First 5 training labels:', y_train[:5])
from sklearn.linear_model import LogisticRegression
lr = LogisticRegression().fit(X_train, y_train)
y_pred_lr = lr.predict(X_test)
print('Number of mislabeled points out of test set of %d points:' % (X_test.shape[0]))
print('Logistic Regression: %d, Score: %f' % ((y_test != y_pred_lr).sum(), lr.score(X_test, y_test)))
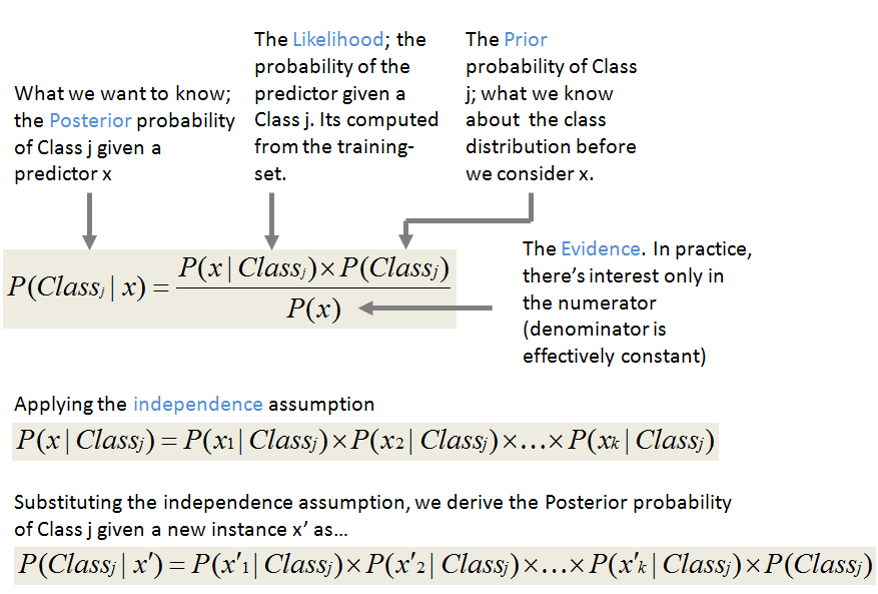
Bayes Theorem¶
- Inputs: independent features
- Outputs: class probabilities
- Bayes Theorem computes the conditional probabilities
Libraries¶
http://scikit-learn.org/stable/modules/naive_bayes.html#naive-bayes
- Gaussian
- Multinomial
- Bernoulli
from sklearn import naive_bayes
# Now that we added noise to our data, we need to scale the features to between [0, 1]
# This is because Naive Bayes cannot handle negative features (throws an error)
from sklearn.preprocessing import MinMaxScaler
min_max_scaler = MinMaxScaler()
min_max_scaler.fit(X_train)
X_train_minmax = min_max_scaler.transform(X_train)
X_test_minmax = min_max_scaler.transform(X_test)
nb_titles = ['Gaussian Naive Bayes', 'Multinomial NB', 'Bernouilli NB']
nb_models = (naive_bayes.GaussianNB(),
naive_bayes.MultinomialNB(),
naive_bayes.BernoulliNB())
nb_models = (model.fit(X_train_minmax, y_train) for model in nb_models)
print('Number of mislabeled points out of test set of %d points:' % (X_test.shape[0]))
for model, title in zip(nb_models, nb_titles):
y_pred = model.predict(X_test_minmax)
wrong = (y_test != y_pred).sum()
score = model.score(X_test_minmax, y_test)
print('%s: %d (score: %.2f)' %(title, wrong, score))
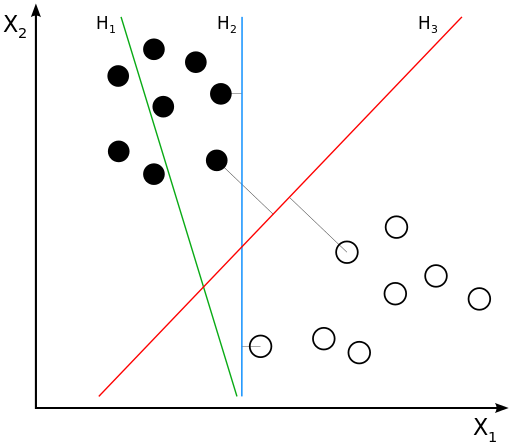
Support Vector Machines¶
- Inputs: features (not necessarily independent)
- Features should be scaled
Output: classes, separated by "hyperplane"
SVM uses "kernel functions" to compute the similarity between input samples
- Find hyperplane with the maximum margin of separation
- Why? Better generalization
Libraries¶
http://scikit-learn.org/stable/modules/generated/sklearn.svm.SVC.html
- Support Vector Classifier
- Different kernel functions to choose from
Scikit-learn has a nifty example that shows how the different kernel functions look like.
To illustrate them, we'll use their code example to train SVM models with only 2 features.
- Why 2 features? Because it's easier to plot in 2-D
# http://scikit-learn.org/stable/auto_examples/svm/plot_iris.html#sphx-glr-auto-examples-svm-plot-iris-py
from sklearn import svm
def make_meshgrid(x, y, h=.02):
"""Create a mesh of points to plot in
Args:
x: data to base x-axis meshgrid on
y: data to base y-axis meshgrid on
h: stepsize for meshgrid, optional
Returns:
xx, yy : ndarray
"""
x_min, x_max = x.min() - 1, x.max() + 1
y_min, y_max = y.min() - 1, y.max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
return xx, yy
def plot_contours(ax, clf, xx, yy, **params):
"""Plot the decision boundaries for a classifier.
Args:
ax: matplotlib axes object
clf: a classifier
xx: meshgrid ndarray
yy: meshgrid ndarray
params: dictionary of params to pass to contourf, optional
"""
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
Z = Z.reshape(xx.shape)
out = ax.contourf(xx, yy, Z, **params)
return out
fig, sub = plt.subplots(nrows=2, ncols=2, figsize=(15, 10))
plt.subplots_adjust(wspace=0.4, hspace=0.4)
# Take the first two features. We could avoid this by using a two-dim dataset
X_train_svc = X_train[:, :2]
# LinearSVC uses liblinear, SVC uses libsvm
# Both are different implementations of SVM
svm_titles = ['LinearSVC (liblinear)',
'SVC (linear kernel)',
'SVC (RBF kernel)',
'SVC (3-degree polynomial kernel)']
# we create an instance of SVM and fit our data. We do not scale our
# data since we want to plot the support vectors
C = 1.0 # SVM regularization parameter
svm_2D_models = (svm.SVC(kernel='linear', C=C),
svm.LinearSVC(C=C),
svm.SVC(kernel='rbf', gamma=0.7, C=C),
svm.SVC(kernel='poly', degree=3, C=C))
svm_2D_models = (clf.fit(X_train_svc, y_train) for clf in svm_2D_models)
X0, X1 = X_train_svc[:, 0], X_train_svc[:, 1]
xx, yy = make_meshgrid(X0, X1)
for clf, title, ax in zip(svm_2D_models, svm_titles, sub.flatten()):
plot_contours(ax, clf, xx, yy,
cmap=plt.cm.coolwarm, alpha=0.8)
ax.scatter(X0, X1, c=y_train, cmap=plt.cm.coolwarm, s=20, edgecolors='k')
ax.set_xlim(xx.min(), xx.max())
ax.set_ylim(yy.min(), yy.max())
ax.set_xlabel('Sepal length')
ax.set_ylabel('Sepal width')
ax.set_xticks(())
ax.set_yticks(())
ax.set_title(title)
plt.show()
Let's compare the performance of SVM with the other Classification models (Logistic Regression, Naive Bayes)
To do that, we retrain the SVM models with the full features.
Exercise - Train and score SVM using different kernels¶
Train SVM models for the 4 kernel functions.
For each model:
- Scale X_train and X_test using sklearn.preprocessing.StandardScaler.
- X_train and X_test are multi-dimensional numpy arrays, so you can pass them directly into the scaler without reshaping.
- Print the number of mislabeled points
- Print the score
Use all the features instead of just the first two.
Which model performs the best?
# Your code here
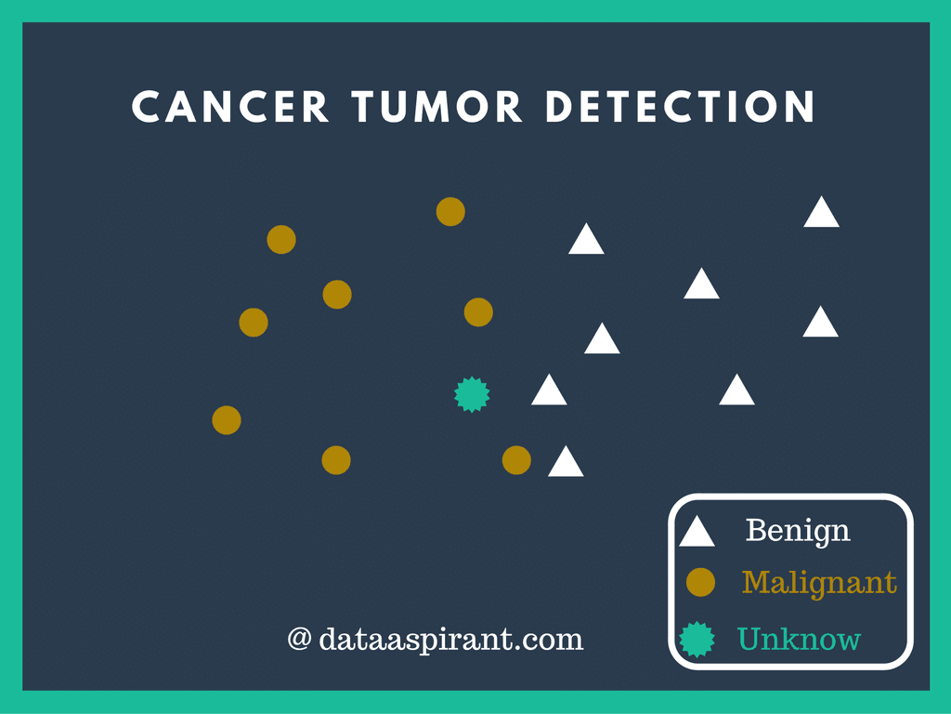
K-nearest Neighbors¶
K-nearest neighbors is a multi-purpose algorithm that can be used for multi-class classification.
- Find K closest neighbors to that sample
- Classify by majority vote of the classes of that sample
(image: dataaspirant.com)
Libraries¶
sklearn.neighbors.KNeighborsClassifier
http://scikit-learn.org/stable/modules/generated/sklearn.neighbors.KNeighborsClassifier.html
from sklearn import neighbors
# K (how many neighbors to consider for the vote)
n_neighbors = [3, 5, 15]
# types of weights
weights = ['uniform', 'distance']
kn_models = []
for k in n_neighbors:
for weight in weights:
model = neighbors.KNeighborsClassifier(k, weights=weight)
model.fit(X_train, y_train)
y_pred = model.predict(X_test)
wrong = (y_test != y_pred).sum()
score = model.score(X_test, y_test)
print('k = %d, weights = %s: %d (score: %.2f)' %(k, weight, wrong, score))
# http://scikit-learn.org/stable/auto_examples/neighbors/plot_classification.html
import matplotlib.pyplot as plt
from matplotlib.colors import ListedColormap
X_train_plot = X_train[:, :2]
h = .02 # step size in the mesh
n_neighbors = 15
# Create color maps
cmap_light = ListedColormap(['#FFAAAA', '#AAFFAA', '#AAAAFF'])
cmap_bold = ListedColormap(['#FF0000', '#00FF00', '#0000FF'])
for weights in ['uniform', 'distance']:
clf = neighbors.KNeighborsClassifier(n_neighbors=n_neighbors, weights=weights)
clf.fit(X_train_plot, y_train)
# Plot the decision boundary. For that, we will assign a color to each
# point in the mesh [x_min, x_max]x[y_min, y_max].
x_min, x_max = X_train_plot[:, 0].min() - 1, X_plot[:, 0].max() + 1
y_min, y_max = X_train_plot[:, 1].min() - 1, X_plot[:, 1].max() + 1
xx, yy = np.meshgrid(np.arange(x_min, x_max, h),
np.arange(y_min, y_max, h))
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
# Put the result into a color plot
Z = Z.reshape(xx.shape)
plt.figure(figsize=(10, 8))
plt.pcolormesh(xx, yy, Z, cmap=cmap_light)
# Plot also the training points
plt.scatter(X_train_plot[:, 0], X_train_plot[:, 1], c=y_train, cmap=cmap_bold,
edgecolor='k', s=20)
plt.xlim(xx.min(), xx.max())
plt.ylim(yy.min(), yy.max())
plt.title("3-Class classification (k = %i, weights = '%s')"
% (n_neighbors, weights))
plt.show()
A Quick Comparison¶
| Criteria | Logistic Regression | Naive Bayes | SVM | K-nearest neighbors |
|---|---|---|---|---|
| Interpretability | Simple | Very simple (Conditional Probabilities) | Hard to understand parameters | Simple, but need to pick K |
| Ease of training | Fast to train | Fast to train | Computationally and memory intensive for high dimensional data | Can be expensive for high dimensional data |
| Requires independent features | Yes, but may still work | Yes, assumes independence | No (don't care) | No (don't care) |
| Feature value ranges | No requirements, scale if vary too widely | No negative features | Must scale to [-1, 1] | No requirements |
| Output usefulness | Returns probabilities and categories | Returns probabilities and categories | Returns probabilities and categories | Returns probabilities and categories |
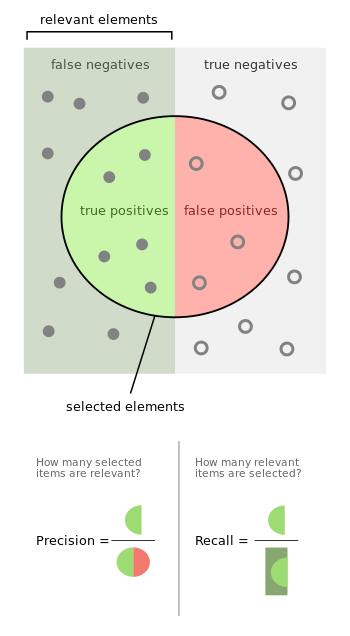
Confusion Matrix¶
| Truth/Prediction | Predicted Happy | Predicted Sad |
|---|---|---|
| Actually Happy | True positive count | False negative count |
| Actually Sad | False positive count | True negative count |
Example¶
| Truth/Prediction | Predicted Happy | Predicted Sad |
|---|---|---|
| Actually Happy | 4 | 3 |
| Actually Sad | 2 | 1 |
Accuracy¶
Meaning: what proportion of the samples did the classifier predict correctly?
$$accuracy(y\_true, y\_pred) = \frac{true\_positives + true\_negatives}{total}$$
What's the accuracy in our example?
Precision¶
Meaning: of the samples the classifier predicted TRUE, what proportion did it get correct (i.e. actually true)? The goal is to reduce false positives.
$$precision(y\_true, y\_pred) = \frac{true\_positives}{true\_positives + false\_positives}$$
What's the precision in our example?
Recall (True Positive Rate / Sensitivity)¶
Meaning: of all the TRUE samples, what proprtion did the classifier predict to be TRUE? (How many of the TRUE predictions did the classifier recall)
$$recall(y\_true, y\_pred) = \frac{true\_positives}{true\_positives + false\_negatives}$$
What's the recall in our example?
Specificity (True Negative Rate)¶
Meaning: of all the FALSE samples, what proportion did the clasifier predict to be FALSE?
$$specificity(y\_true, y\_pred) = \frac{true\_negatives}{true\_negatives + false\_positives}$$
What's the specificity in our example?
F1 score / F measure¶
Meaning: a combination of both Precision and Recall in 1 number. This is a harmonic mean of two numbers that uses the standard equation 2AB/(A+B).
$$F(y\_true, y\_pred) = 2 * \frac{precision * recall}{precision + recall}$$
Libraries¶
http://scikit-learn.org/stable/modules/model_evaluation.html#classification-metrics
- classification_report
- confusion_matrix
- accuracy_score
- f1_score
- ...
from sklearn.metrics import classification_report, confusion_matrix
print('Logistic Regression:')
print(classification_report(y_test, y_pred_lr))
cm = confusion_matrix(y_test, y_pred_lr)
print(cm)
# Colormaps:
# https://matplotlib.org/gallery/color/colormap_reference.html
plt.matshow(cm, cmap='Blues')
plt.colorbar()
iris.target_names # the integer values [0, 1, 2] map to these labels
Exercise - Evaluation Metrics for Naive Bayes and SVM Classifiers¶
For the Naive Bayes and SVM models we've seen so far:
- Get the classification metrics.
- Plot the confusion matrix
- How would you interpret the results?
# Get the classification metrics
# Your code here
# Plot the confusion matrices for Naive Bayes
# Your code here
# Plot the confusion matrices for SVM
# Your code here
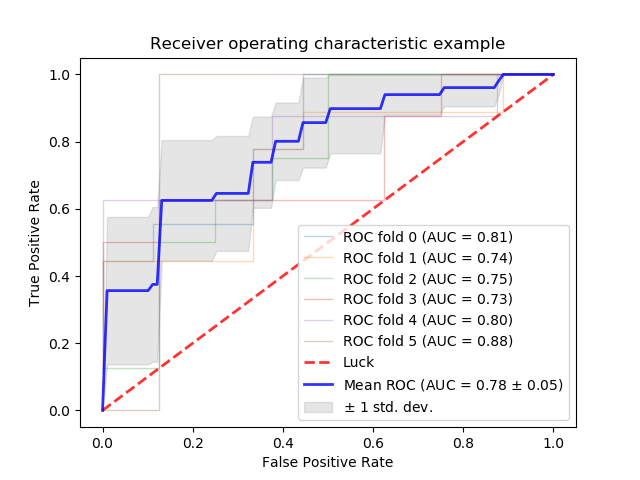
Area under ROC curve¶
ROC curve: Receiver Operating Characteristic
A plot of True Positive Rate (Recall) vs. False Positive Rate (1-Specificity)
Larger area under ROC curve: better performance
Libraries¶
sklearn.metrics.roc_curve: http://scikit-learn.org/stable/modules/generated/sklearn.metrics.roc_curve.html
sklearn.metrics.auc: http://scikit-learn.org/stable/modules/generated/sklearn.metrics.auc.html
ROC curves and Multi-class Classification¶
- ROC is typically for binary classification
- To plot ROC for multi-class:
- Either draw 1 curve per class
- Or compute average for all the classes
# http://scikit-learn.org/stable/auto_examples/model_selection/plot_roc.html
from sklearn.multiclass import OneVsRestClassifier
from sklearn.metrics import roc_curve, auc
from sklearn.preprocessing import label_binarize
iris = datasets.load_iris()
X = iris.data
# binarize the y labels
y = label_binarize(iris.target, classes=[0, 1, 2])
n_classes = y.shape[1]
print(y)
# Add noisy features to make the problem harder
random_state = np.random.RandomState(0)
n_samples, n_features = X.shape
X = np.c_[X, random_state.randn(n_samples, 200 * n_features)]
# Shuffle and split train/test
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=.5,
random_state=42)
classifier = OneVsRestClassifier(LogisticRegression())
y_score = classifier.fit(X_train, y_train).decision_function(X_test)
y_score
# Compute ROC curve and AROC for each class
fpr = dict()
tpr = dict()
roc_auc = dict()
for i in range(n_classes):
fpr[i], tpr[i], _ = roc_curve(y_test[:, i], y_score[:, i])
roc_auc[i] = auc(fpr[i], tpr[i])
# Plot the ROC curves
fig, ax = plt.subplots(figsize=(15, 10))
for i in range(n_classes):
ax.plot(fpr[i], tpr[i], label='ROC curve of class %d (area = %0.2f)' % (i, roc_auc[i]))
ax.legend()